Week 7: Starting out
EA Enterobacter aerogenes -bacilli
EC Escherichia coli -bacilli
PA Pseudomonas aeruginosa -bacilli
PS Providencia stuartii -bacilli
SM Serratia marcescens -bacilli
BS Bacillus subtilis +bacilli
CX Corynebacteria xerosis +bacilli
BS Bacillus cereus +bacilli
SE Staphylococcus epidermidis +cocci
SA Staphylococcus aureus +cocci
EF Enterococcus faecalis +cocci
I waited 24 hours for the samples to develop, and half turned out quite well. On the samples that were good to use, there were distinguishable single colonies of bacteria that we could take our first pictures of. However, the other half needed to be redone as my streaks were heavy-handed. Not bad for my first time working with bacteria, though! After redoing the samples, Joshua and I used a dissecting microscope. However, the pictures needed to be more detailed to show distinct characteristics of the colony other than it being a sphere (at least for our SA sample). Therefore, we switched to a regular microscope, where the pictures turned out much better! We will likely use that microscope for the rest of our samples. On this day, we did 3 samples. One was E. Coli, which had a very irregular shape, but the other two bacteria we scanned (SA and EF). These two looked very similar, reddish-brown spheres with a darker center. Therefore, we plan to make 3 different algorithms for each class so that the program can distinguish between class types instead of looking for characteristics in all 8 samples simultaneously. This will be our plan until we figure out how to take more detailed photos.
For now, my next few weeks will look similar as we will need to take many, many photos to create a reliable algorithm. We made significant progress to start, though!
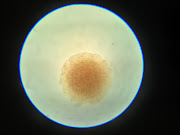
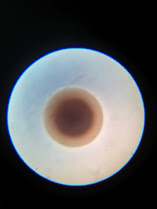




Hi Maya, this is also my first week with TRAIN program. I think it would be cool to be able to identify bacteria from a picture. Working with bacteria can be difficult but it's good you have an open and positives mind set to it. What made you choose this experiment? Good Luck!
ReplyDelete